
Research
How do species traits link to biodiversity through time and space?
From phylogenetic inference to comparative methods
I am broadly interested in how biodiversity arises and is maintained (or not) in an ever changing environment. To look at this I work on species traits at both macroevolutionary and macroecological levels through time and space. The bulk of my work consists in developing or improving models to look at species traits in phylogenetic inferences all the way to comparative methods.
Traits diversity (disparity!) through time and space


There has been a recent an exciting shift in ecology and evolution to focus work more on species traits rather than the raw species numbers. Looking at species traits allows to better understand how species interact with each other and how they are affected by environmental changes whether these are the current anthropogenic ones or biotic and abiotic changes in the deep past. To do that, researchers have been working on concepts such as traits dissimilarity in ecology and traits disparity in palaeontology (which I would argue are the same). I am interested in how to bridge the gap between these two concepts and how using them on modern and fossil data can tell us on the state of biodiversity through time and space.
Morphological characters in phylogenetics


To study biodiversity changes in deep time (and in the present!) we need to link fossil and living species in the same phylogenies. Our understanding of molecular evolution has remarkably enhanced in the last two decades leading to excellent knowledge in both the mechanisms underlying DNA evolution as well as the software implementations to compute it efficiently. Unfortunately, our understanding of more complex morphological character evolution is still lacking and require both theoretical and practical improvements. I am working on how to link these two methods and I am really interested in the challenges that comes with it.
Macroecology
Although a lot of my work focuses on macroevolution, I am also working actively on using the methods I develop to answer macroecological quesitions. By focusing on traits rather than number of species we can answer a lot of very specific questions in macroecology and see and understand more generally how species interact with their environment.
Didn't understood a word?
Read my Up-Goer 5 research:
I am interested in looking how animals forms changed through time. I use computers to make not real (but real looking!) changes of animals forms through time to understand how the things (like the death of many animals or when a group of animal try to fight another one) can change the forms of the animals or even allow new animal forms to appear. This also allows me to look if groups of animals are in the same family and how their family changed through time.
Academic Career
- 2023-present
- Independent NERC fellow at the University of Sheffield - UK
- 2020-2023
- Research Associate at the University of Sheffield - UK
- Working in Gavin Thomas's and Andrew Beckerman's and Natalie Cooper's groups.
- 2017-2019
- Postdoctoral Research Fellow at the University of Queensland, Brisbane - Australia
- Working in Vera Weisbecker's and Matt Phillips's groups.
- 2016-2017
- Research Associate at Silwood Park Campus, Imperial College London - UK
- Working in Martin Brazeau's group.
Education
- 2012-2015
- PhD in Zoology at Trinity College Dublin - IE
- Examined by Prof Philip Donoghue (University of Bristol) and Dr Trevor Hodkinson (Trinity College Dublin). Awarded as it stands (no corrections).
- Macroevolution with living and fossil species
- Examined by Prof Philip Donoghue (University of Bristol) and Dr Trevor Hodkinson (Trinity College Dublin). Awarded as it stands (no corrections).
- 2010-2012
- Master degree in Palaeontology at the Université de Montpellier - FR
- with honours, ranking 3/13
- 2nd year project: Molecular clocks and evolutionary rates among primates: a palaeontological and genomic approach.
- 1st year project: Southern France Upper Cretaceous theropod teeth variability.
- with honours, ranking 3/13
- 2007-2010
- Bachelor degree in Biology at the Université de Montpellier - FR
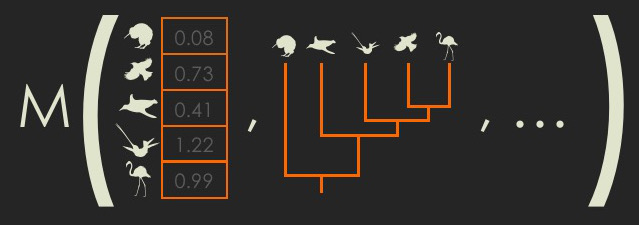
 What is disparity?
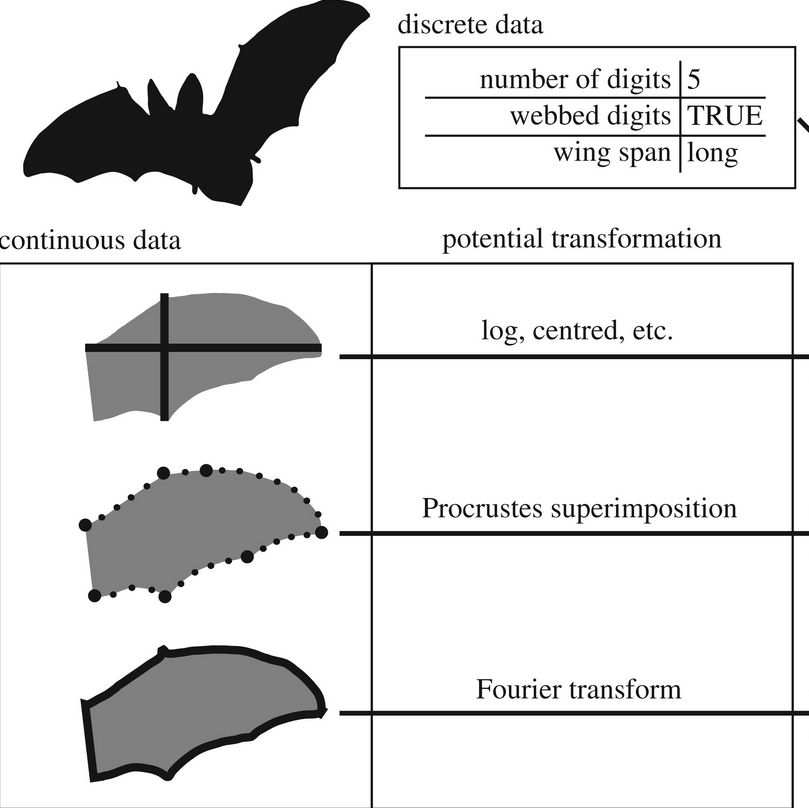
What is disparity? Measuring disparity
Measuring disparity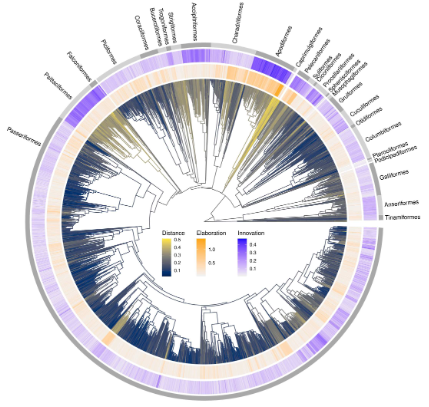 Elaboration and innovation
Elaboration and innovation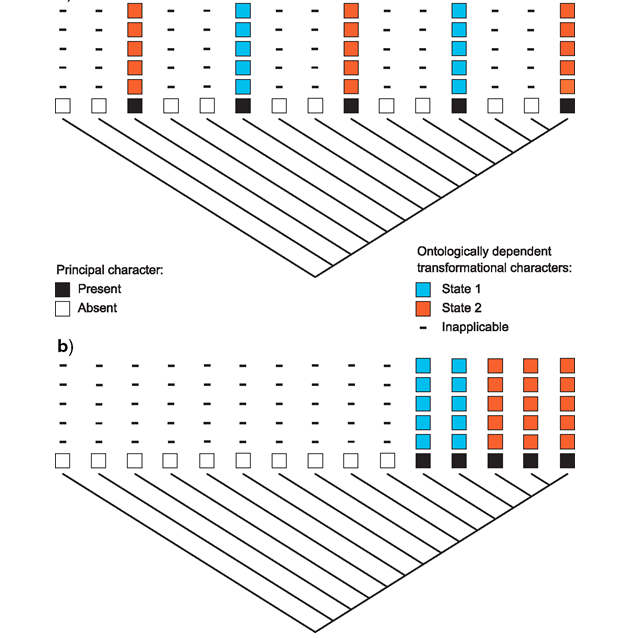 Inapplicable data in phylogenetics
Inapplicable data in phylogenetics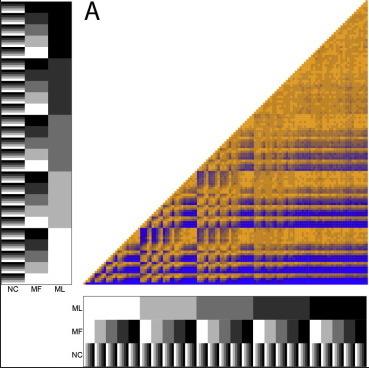 Missing data in Total Evidence
Missing data in Total Evidence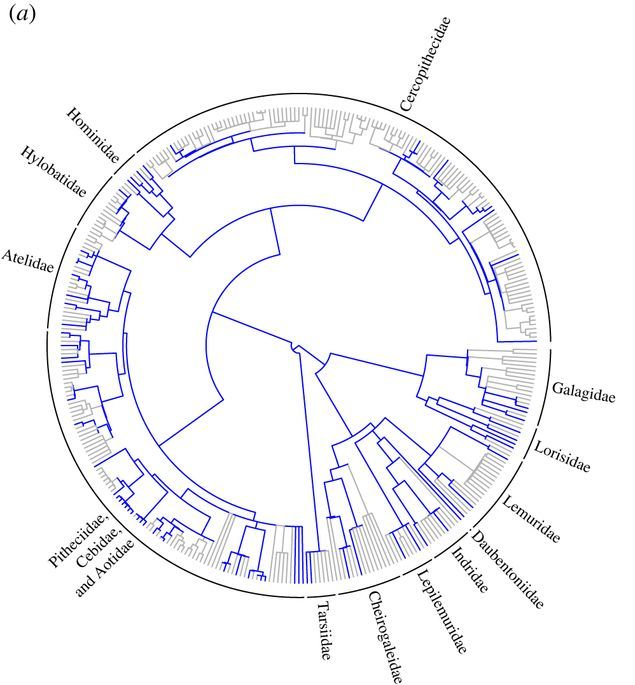 Character correlation in phylogenetics
Character correlation in phylogenetics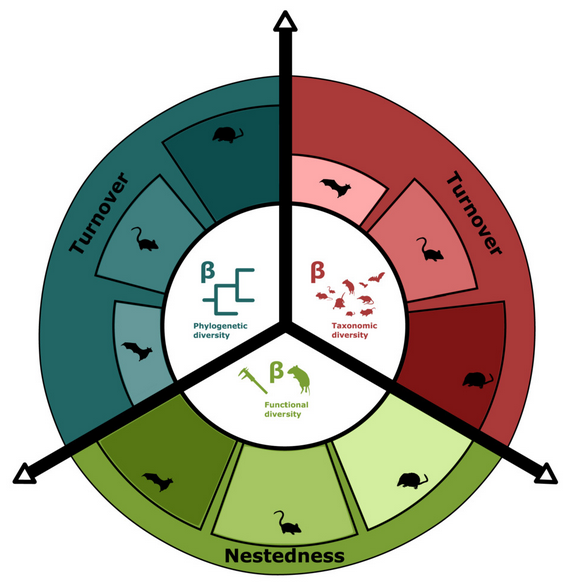 macroecology at continental scales
macroecology at continental scales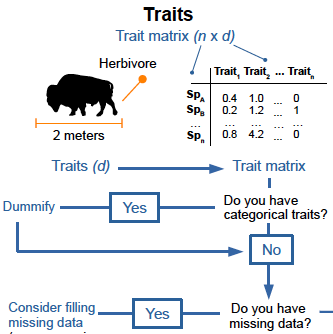 functional ecology
functional ecology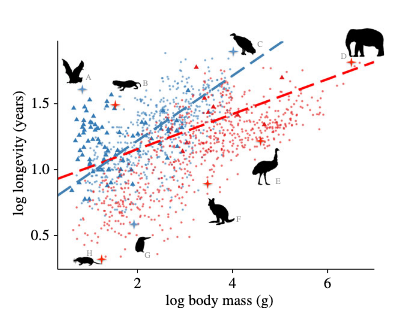 macroecology of longevity
macroecology of longevity