Publications
You can check out my cynical academic game scores (h-index, number of citations, etc.) on my Google Scholar page and other academic vanity metrics on the various links at the bottom of my my main page (Publons, ResearchID, ResearchGate and other commercial website that like diverting public money).
I am aiming to publish all my work as Open Access, however, it sometimes happen that I just have no funding to do so or that the main authors of the papers I collaborated on cannot make their work Open Access either (for their own reasons).
Therefore, you can find a link to the free versions for each paper below by clicking on the Open Access or the PDF logo. This was not specifically approved my co-authors (although I believe they don't mind) nor by the publishers (and I'm sure they do mind).
25- The Greatest Extinction Event in 66 Million Years? Contextualising Anthropogenic Extinctions.
Hatfield J, Allen B, Carroll T, Dean C, Deng S, Gordon J, Guillerme T, Hansford J, Hoyal Cuthill J, Mannion P, Martins I, Payne A, Shipley A, Thomas C, Thompson J, Woods L, Davis K. Global Change Biology 2025 https://doi.org/110.1111/gcb.70476.
-
Key words: Anthropocene, biodiversity, Cenozoic, extinction, mass extinction, sixth mass extinction.
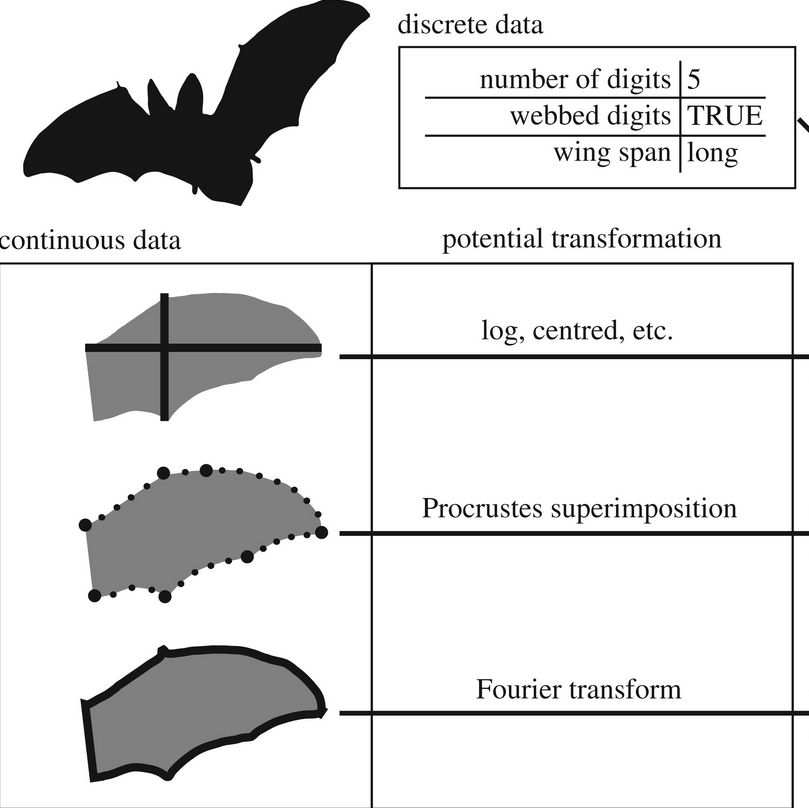
24- The what, how and why of trait-based analyses in ecology.
Guillerme T, Cardoso P, Jørgensen MW, Mammola, S, Matthews TJ. Ecography 2025 - Editor's choice - https://doi.org/10.1111/ecog.07580.
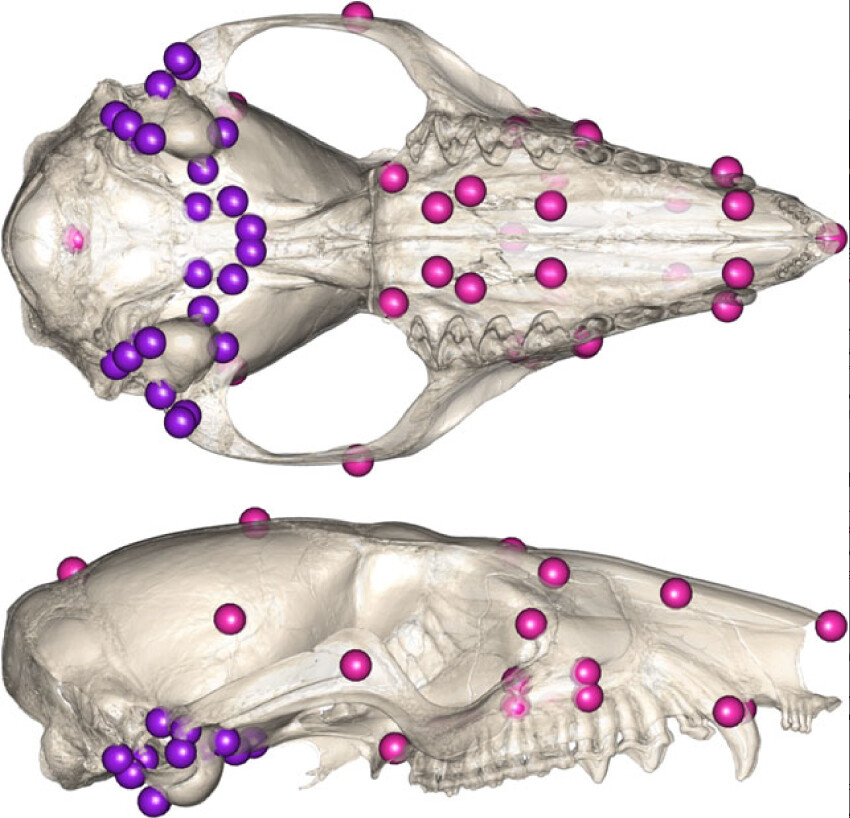
23- Beyond CREA: evolutionary patterns of non-allometric shape variation and divergence in a highly allometric clade of murine rodents.
Marcy AE, Mitchell DR. Guillerme T, Phillips MJ, Weisbecker V. Ecology and Evolution 2024 doi:10.1002/ece3.11588.

-
Key words: allometry, CREA, geometric morphometrics, integration, modularity, Muridae, stabilising selection.
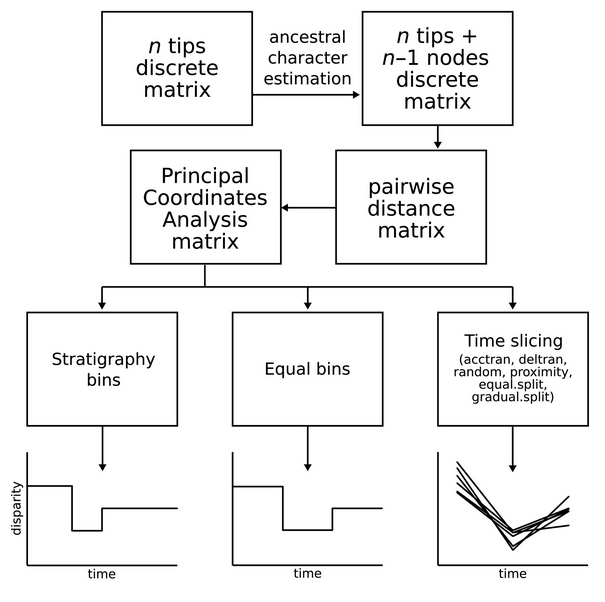
22- Calculating functional diversity metrics using neighbor-joining trees.
Cardoso P, Guillerme T, Mammola S, Matthews TJ, Rigal F, Graco-Roza C, Stahls G, Carvalho JC. Ecography 2024 doi.org/10.1111/ecog.07156.
-
Key words: Convex hulls, dendrograms, functional divergence, functional diversity, functional regularity, functional traits, hypervolumes, neighbor-joining.
21- treats: a modular R package for simulating trees and traits.
Guillerme T. Methods in Ecology and Evolution 2024 doi.org/10.1111/2041-210X.14306.
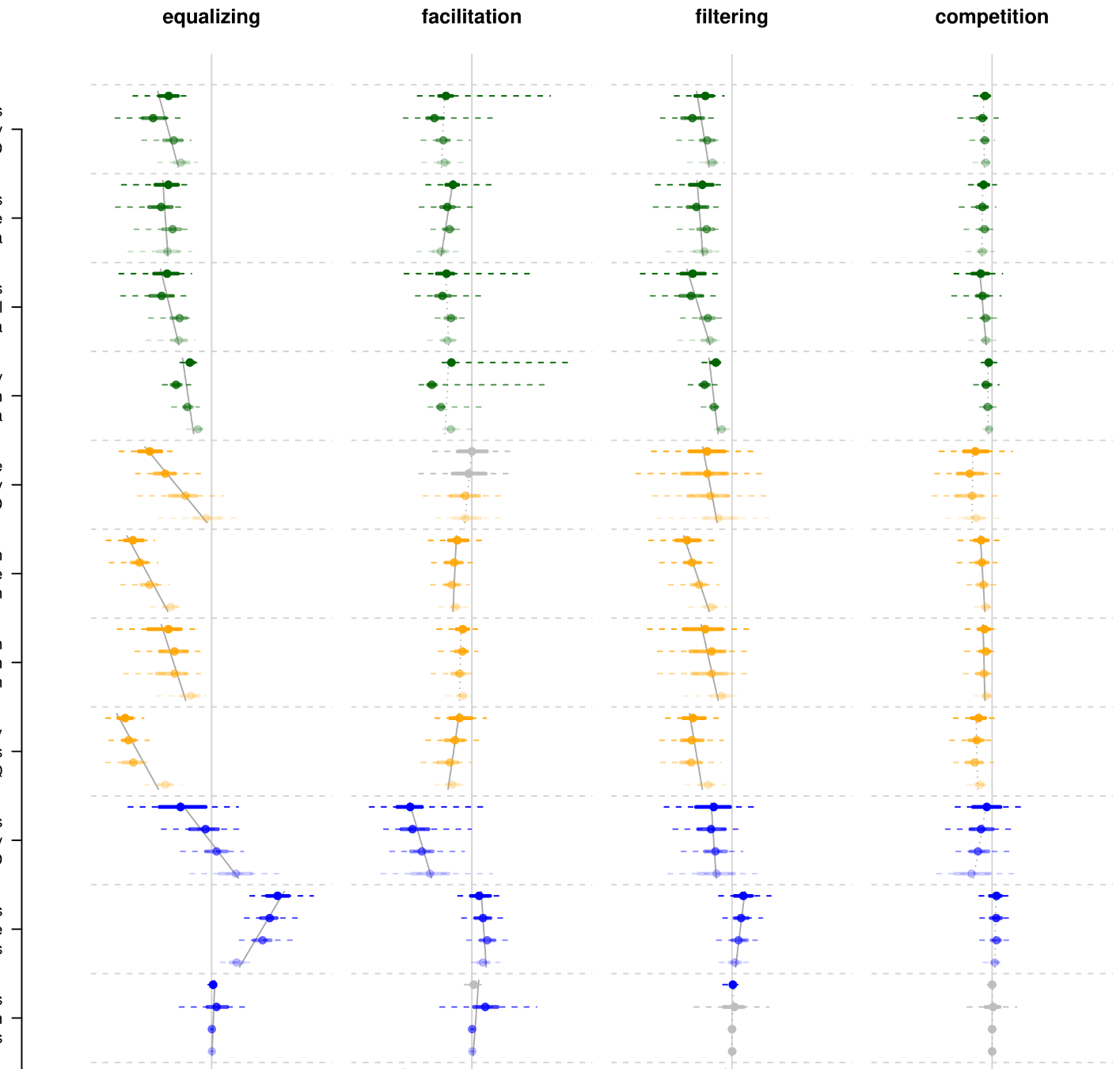
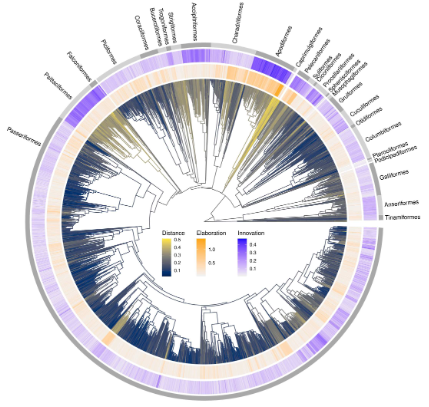
20- Innovation and elaboration on the avian tree of life. Guillerme T, Bright JA, Cooney CR, Hughes EC, Varley ZK, Cooper N, Beckerman AP, Thomas GH. Science Advances 2023 doi.org/10.1126/sciadv.adg1641.
19- Multiple modes of inference reveal less phylogenetic signal in marsupial basicranial shape compared with the rest of the cranium.Weisbecker V, Beck RMD, Guillerme T, Harrington AR, Lange-Hodgson L, Lee MSY, Mardon K, Phillips MJ. Philosophical Transactions of the Royal Society B 2023 doi.org/10.1098/rstb.2022.0085.
18- Dispersal ability is associated with contrasting patterns of beta diversity of African small mammal communities.Monadjem A, Healy K, Guillerme T, Kane A. Journal of Biogeography 2023 doi.org/10.1111/jbi.14532.
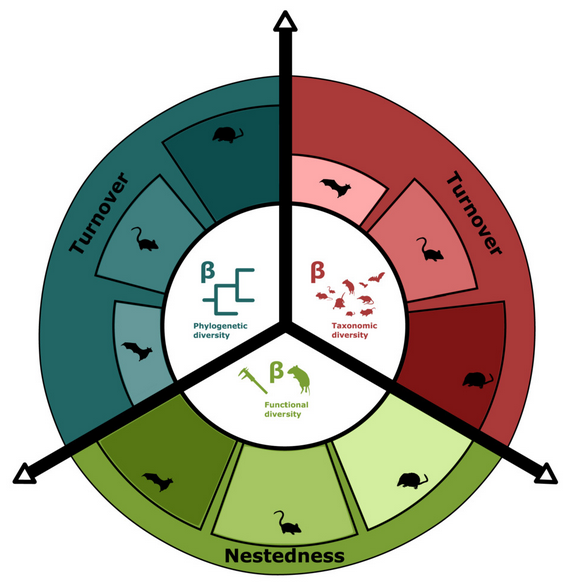
-
Key words: alpha diversity, beta diversity, biological trait, disparity, evenness, functional dendrogram, functional dispersion functional regularity, functional richness, hypervolume
17- Concepts and applications in functional diversity.Mammola S, Carmona CP, Guillerme T, Cardoso P. Functional Ecology 2021 doi.org/10.1111/1365-2435.13882.
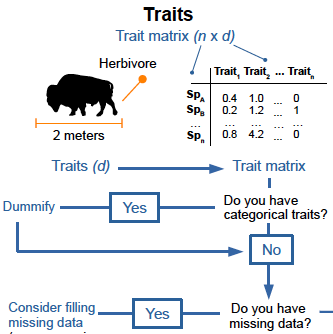
-
Key words: alpha diversity, beta diversity, biological trait, disparity, evenness, functional dendrogram, functional dispersion functional regularity, functional richness, hypervolume
16- Endochondral bone in an Early Devonian 'placoderm' from Mongolia.
Brazeau MD, Giles S, Dearden RP, Jerve A, Ariunchimeg Y, Zorig E, Guillerme T, Castiello M. Nature Ecology and Evolution 2020 doi.org/10.1038/s41559-020-01290-2.
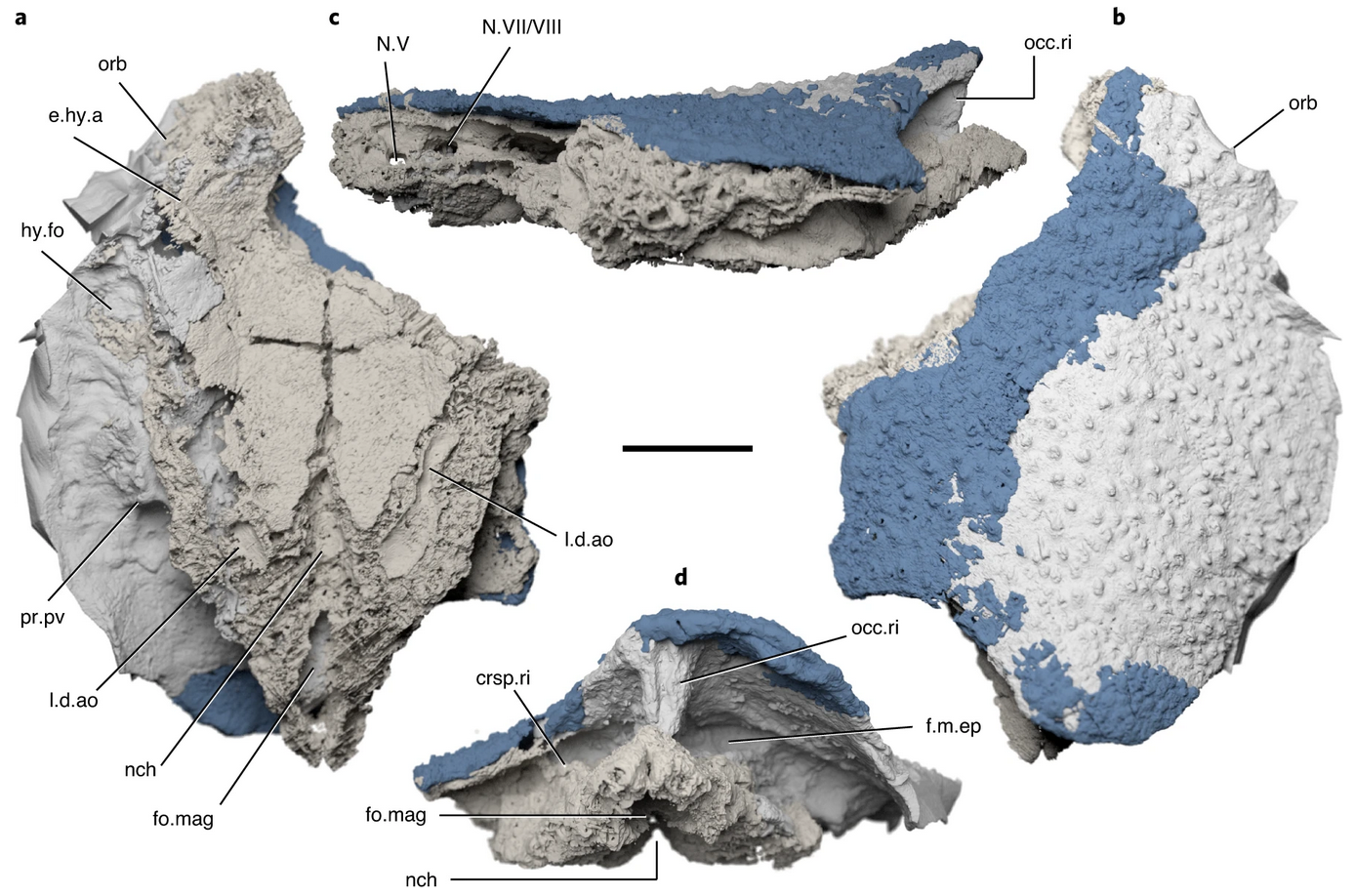
-
Key words: Minjinia, placoderm, Endochondral bone, devonian.
Personal contribution: run the ancestral states estimation analysis and helped with writing the paper.
15- Australian rodents reveal conserved Cranial Evolutionary Allometry across 10 million years of murid evolution.
Marcy AE, Guillerme T, Sherratt E, Rowe KC, Phillips MJ, Weisbecker V. American Naturalist 2020 doi.org/10.1086/711398.
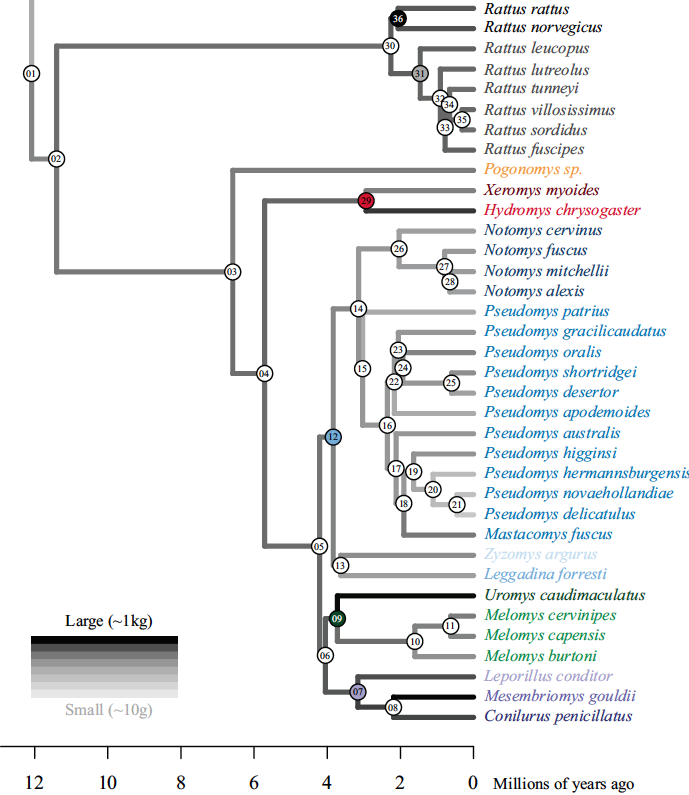
-
Key words: allometric facilitation, CREA, geometric morphometrics, molecular phylogeny, Murinae, stabilizing selection.
Personal contribution: helped developing the methods, analysing the data and writing the paper.
14- Skull shape of a widely distributed, endangered marsupial reveals little evidence of local adaptation between fragmented populations.
Viacava P, Blomberg SP, Sansalone G, Phillips MJ, Guillerme T, Cameron SF, Wilson RS, Weisbecker V. Ecology and Evolution 2020 doi.org/10.1002/ece3.6593.
-
Key words: conservation, Dasyurus hallucatus, geometric morphometrics, intraspecific variation, procrustes ANOVA, shape variation, variation partitioning.
Personal contribution: helped with designing the experiment, running the analyses and with writing the paper. Blog post by the main author.
13- The complex effects of mass extinctions on morphological disparity.
Puttick MN, Guillerme T, Wills MA. Evolution 2020 doi:10.1111/evo.14078.
-
Key words: disparity, mass extinctions, phylogenetic comparative methods, traits, macroevolution.
Personal contribution: helped with designing the experiment and with writing the paper.
12- Shifting spaces: which disparity or dissimilarity measurement best summarise occupancy in multidimensional spaces?
Guillerme T, Puttick MN, Marcy AE, Weisbecker V. Ecology and Evolution 2020 doi:10.1002/ece3.6452.
11- Disparities in the analysis of morphological disparity.
Guillerme T, Cooper N, Brusatte SL, Davis KE, Jackson AL, Gerber S, Goswami A, Healy K, Hopkins MJ, Jones MEH, Lloyd GT, O’Reilly JE, Pate A, Puttick MN, Rayfield E, Saupe EE, Sherratt E, Slater GJ, Weisbecker V, Thomas GH, Donoghue PCJ. Biology Letters 2020 doi:10.1098/rsbl.2020.0199.
10- Individual variation of the masticatory system dominates 3D skull shape in the herbivory-adapted marsupial wombats.
Weisbecker V* , Guillerme T* , Speck C, Sherratt E, Mehari Abraha H, Sharp A, Terhune CE, Collins S, Johnston S, Panagiotopoulou O*. Frontiers in Zoology 2019 doi:10.1186/s12983-019-0338-5.
-
* Corresponding authors.
Key words: Marsupial, mastication, constraint, cranium, mandible, geometric morphometrics.
Personal contribution: developed the method and helped writing the manuscript.
9- An algorithm for morphological phylogenetic analysis with inapplicable data.
Brazeau MD*,Guillerme T*, Smith MR*. Systematic Biology 2019 doi:10.1093/sysbio/syy083.
-
* All authors contributed equally.
Key words: cladistic analysis, inapplicable data, character independence, phylogenetic tree search, character optimization.
8- dispRity: a modular R package for measuring disparity.
Guillerme T. Methods in Ecology and Evolution 2018 doi:10.1111/2041-210X.13022.
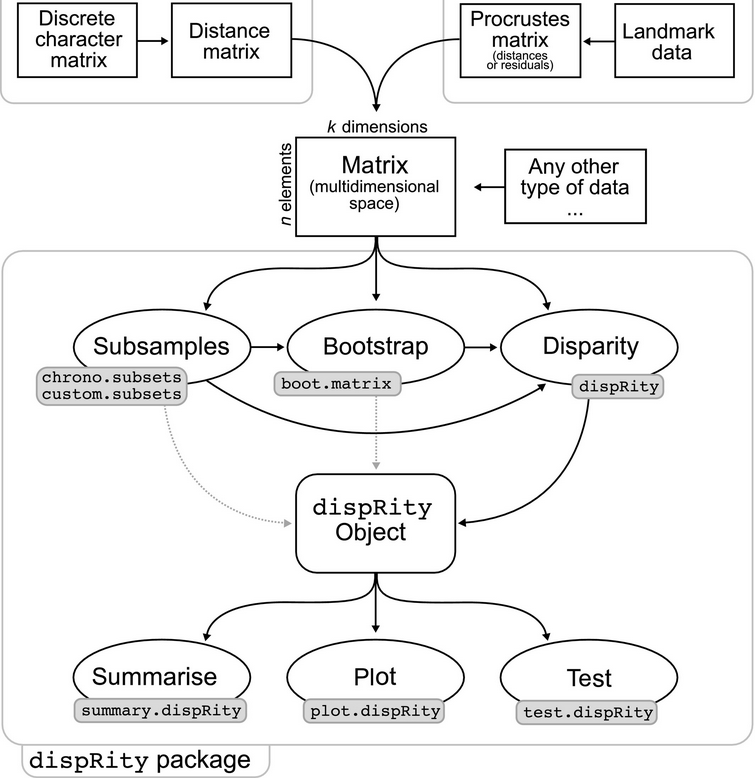
-
Key words: disparity, ordination, multidimensionality, disparity-through-time, palaeobiology, ecology.
Part of a Virtual Issue in Methods in Ecology and Evolution: Phylogenetics and Comparative Methods: The Dark and Bright Sides.
7- Time for a rethink: time sub-sampling methods in disparity-through-time analyses.
Guillerme T, Cooper N. Palaeontology 2018 doi:10.1111/pala.12364.
6- SIDER: An R package for predicting trophic discrimination factors of consumers based on their ecology and phylogenetic relatedness.
Healy K, Guillerme T, Kelly S, Inger R, Bearhop S, Jackson A Ecography 2018 doi:10.1111/ecog.03371
-
Key words: SIDER Trophic Enrichment Factors, Comparative Analysis, Stable Isotope Analysis.
Personal contribution: co-developed the R package.
5- Quaternary vertebrate faunas from Sumba, Indonesia: implications for Wallacean biogeography and evolution.
Turvey S, Crees JJ, Hansford J, Jeffree TE, Crumpton N, Kurniawan I, Setiyabudi E, Guillerme T, Paranggarimu U, Dosseto A, van den Bergh GD. Proceedings of the Royal Society of London B 2017 doi:10.1098/rspb.2017.1278
-
Key words: biogeography, island evolution, murid, Quaternary extinctions, Stegodon, Wallacea.. Personal contribution: ran the phylogenetic analysis.
4- A Recipe for Scavenging - the natural history of a behaviour.
Kane A, Healy K, Guillerme T, Ruxton G, Jackson AL Ecography 2016 doi:10.1111/ecog.02817.
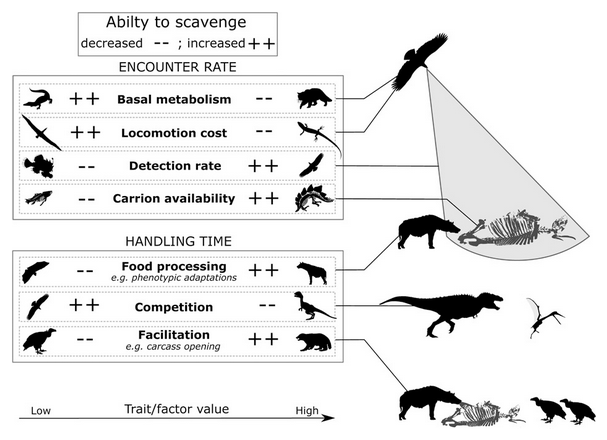
-
Key words: scavenging; carrion; vertebrates.Personal contribution: helping designing and writing the structure of the review. This paper was featured as the Ecography cover for February 2017. See associated blog post.
3- Assessment of available anatomical characters for linking living mammals to fossil taxa in phylogenetic analyses
Guillerme T, Cooper N. Biology Letters 2016 doi:10.1098/rsbl.2015.1003.
-
Key words: Total Evidence Method, phylogenetic clustering, extinct, topology, discrete anatomical matrix. This was an invited submission for a special issue: Putting fossils in trees. Presentation (08/2015) ; Blog post.
2- Effects of missing data on topological inference using a Total Evidence approach.
Guillerme T, Cooper N. Molecular Phylogenetics and Evolution 2016 doi:10.1016/j.ympev.2015.08.023
.
-
Key words: Morphological characters, Bayesian, Maximum Likelihood, topology, fossil, living. Presentation (08/2015) ; Blog post.
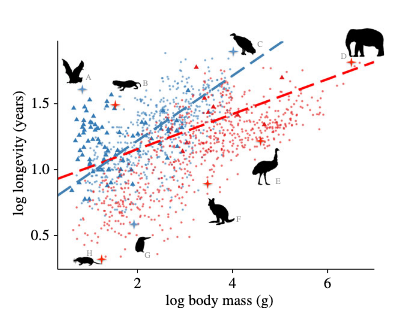
1- Ecology and mode of life explain lifespan variation in birds and mammals.
Healy K, Guillerme T, Finlay S, Kane A, Kelly SBA, McClean D, Kelly DJ, Donohue I, Jackson AL, Cooper N. Proceedings of the Royal Society of London B. 2014 doi:10.1098/rspb.2014.0298
.
-
Key words: Longevity, extrinsic mortality, MCMCglmm, volant, non-volant. Personal contribution: collecting the data, writing the code and running the analysis. Blog post: about the paper and the collaborative project.
dispRity
For easily measuring disparity and or dissimilarity in R with a modular architecture allowing to use any metric you want!
Guillerme T dispRity: a modular R package for measuring disparity. Methods in Ecology and Evolution. doi:10.1111/2041-210X.13022.
treats
For easily simulating any kind of tree and some associated traits. You can also add any event you want to your simulation, like mass extinctions!
Guillerme T treats: a modular R package for simulating trees and traits. Methods in Ecology and Evolution. doi:10.1111/2041-210X.14306.
moms
For visualising and testing disparity or dissimilarity metrics
Guillerme T, Puttick MN, Marcy AE, Weisbecker V.Shifting spaces: which disparity or dissimilarity measurement best summarise occupancy in multidimensional spaces? Ecology and Evolution doi:10.1002/ece3.6452.
landvR
For playing around with landmarks from diverse morphometric geometric packages
Guillerme T & Weisbecker V. landvR: tools for measuring landmark position variation. ZENODO. 10.5281/zenodo.2620785.
Inapp + morphylib
For counting inapplicable characters in discrete morphological phylogenies
Brazeau MD, Smith MR, Guillerme T. morphylib: a C library for calculating tree length under parsimony with inapplicable data. ZENODO. 10.5281/zenodo.815372.
mulTree
For running the MCMCglmm package on a tree distribution
Guillerme T & Healy K. mulTree: a package for running MCMCglmm analysis on multiple trees. ZENODO. 10.5281/zenodo.12902.
mcmcmcglmmm
For running the MCMCglmm package on a tree distribution but for very large datasets
Guillerme T, Bright JA, Cooney CR, Hughes EC, Varley ZK, Cooper N, Beckerman AP, Thomas GH. Innovation and elaboration on the avian tree of life. Science Advances. doi:10.1126/sciadv.adg1641.
SIDER
For estimating stable isotopes using phylogenies